Part:BBa_K2715026
Constitutive clostridial promoter Pcsp_fdx (BBa_K2715011), strong RBS and GusA reporter
Usage and Biology
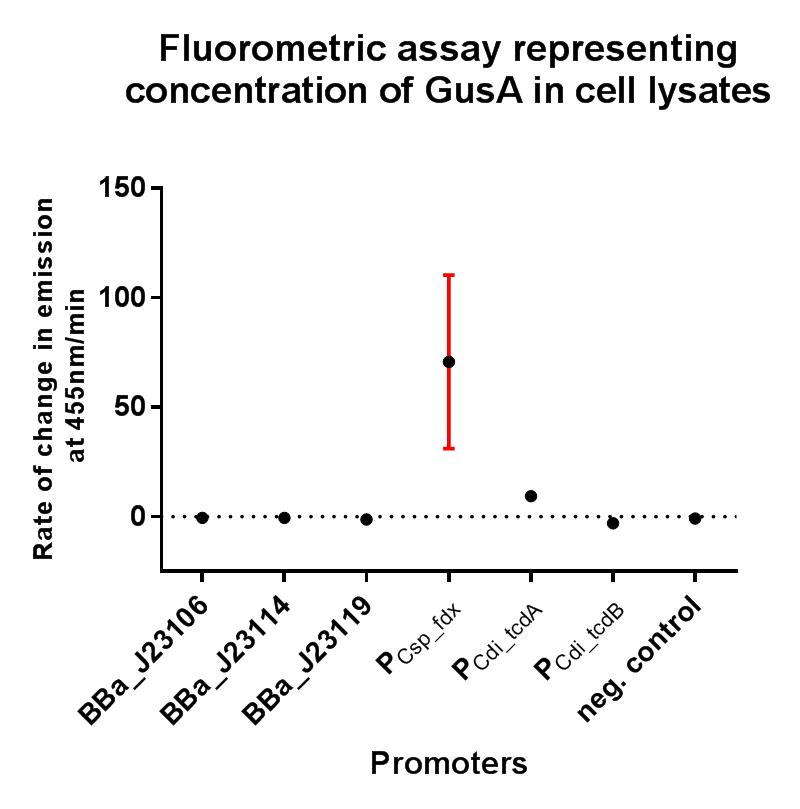
Our project required the use of strong constitutive promoters which would function well in the non-model Gram-positive organism Clostridium difficile. We also wanted to establish whether these promoters functioned in E. coli, as this could have implications for cloning stages and vector assembly when trying to build constructs containing potentially toxic genes. In order to put the strength of these promoters into context, we decided upon a GFP fluorescence assay in E. coli using the iGEM Interlab calibration curves, and also to compare their strengths to the Interlab positive and negative controls. For characterisation in C. difficile we decided to use the reporter gene gusA. The GFP protein requires oxygen in order to fluoresce, so it isn’t suitable as a reporter gene in this context. The Β-Glucuronidase encoded by gusA functions as a very sensitive and specific reporter gene in anaerobic organisms. A substrate for the Β-Glucuronidase, 4-methylumbelliferyl glucuronide (4-MUG), can be added to the cell lysate immediately before the assay, and the production of the fluorescent compound 4-methylumbelliferone (4-MU) measured over time in a spectrophotometer.
This promoter was taken from the related species Clostridium sporogenes, driving expression of the ferredoxin gene, which has been shown in previous research to generally have strong expression in clostridial species.
Characterisation
In this composite part we've added a strong RBS BBa_K2715009, shown to function in Gram-positive and Gram-negative organisms, downstream of the BBa_K2715011 clostridial promoter, driving expression of gusA taken from BBa_K330002. The construct is part of a family of composite parts which all share the same strong RBS and gusA gene and were all characterised in the same plasmid backbone and in parallel in a fluorescence assay, the results of which can be seen below. The negative control in this case is the empty modular vector pMTL84151 which showed no fluorescence.
The gusA containing composites used to assay the promoter activities in C. difficile are listed below.
BBa_K2715025
BBa_K2715026
BBa_K2715027
BBa_K2715028
BBa_K2715029
BBa_K2715030
BBa_K2715031
The plasmid used for this characterisation in C. difficile is displayed below.
Conclusions
This composite part has been used to characterise the expression of K2715011 when used in conjugation with a strong RBS shown to function in both Gram-positive and Gram-negative organisms, and its strength can be quantified in E. coli using the iGEM 2018 interlab units of fluorescence with the composite part K2715002, and in C. difficile using this composite part described here when performing a GUS assay. The expression that was observed from this ferredoxin promoter was greater than all other tested promoters, demonstrating its suitability for applications where a high rate of expression is required in both non-model Gram-positive and Gram-negative organisms.
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
Heap, J.T., Pennington, O.J., Cartman, S.T. and Minton, N.P., 2009. A modular system for Clostridium shuttle plasmids. Journal of microbiological methods, 78(1), pp.79-85.
Davis, D.F., Ward, W.W. and Cutler, M.W., 1994. Posttranslational chromophore formation in recombinant GFP from E. coli requires oxygen. In Bioluminescence and Chemiluminescence: Fundamentals and Applied Aspects. Proceedings of the 8th International Symposium on Bioluminescence and Chemiluminescence, Cambridge. Wiley, New York, NY (pp. 569-599).
Chiu, N.H. and Watson, A.L., 2017. Measuring β‐Galactosidase Activity in Gram‐Positive Bacteria Using a Whole‐Cell Assay with MUG as a Fluorescent Reporter. Current protocols in toxicology, 74(1), pp.4-44.
| None |